Overview
Chronic kidney disease (CKD) affects ∼10% of the population worldwide and ultimately can lead to kidney failure.
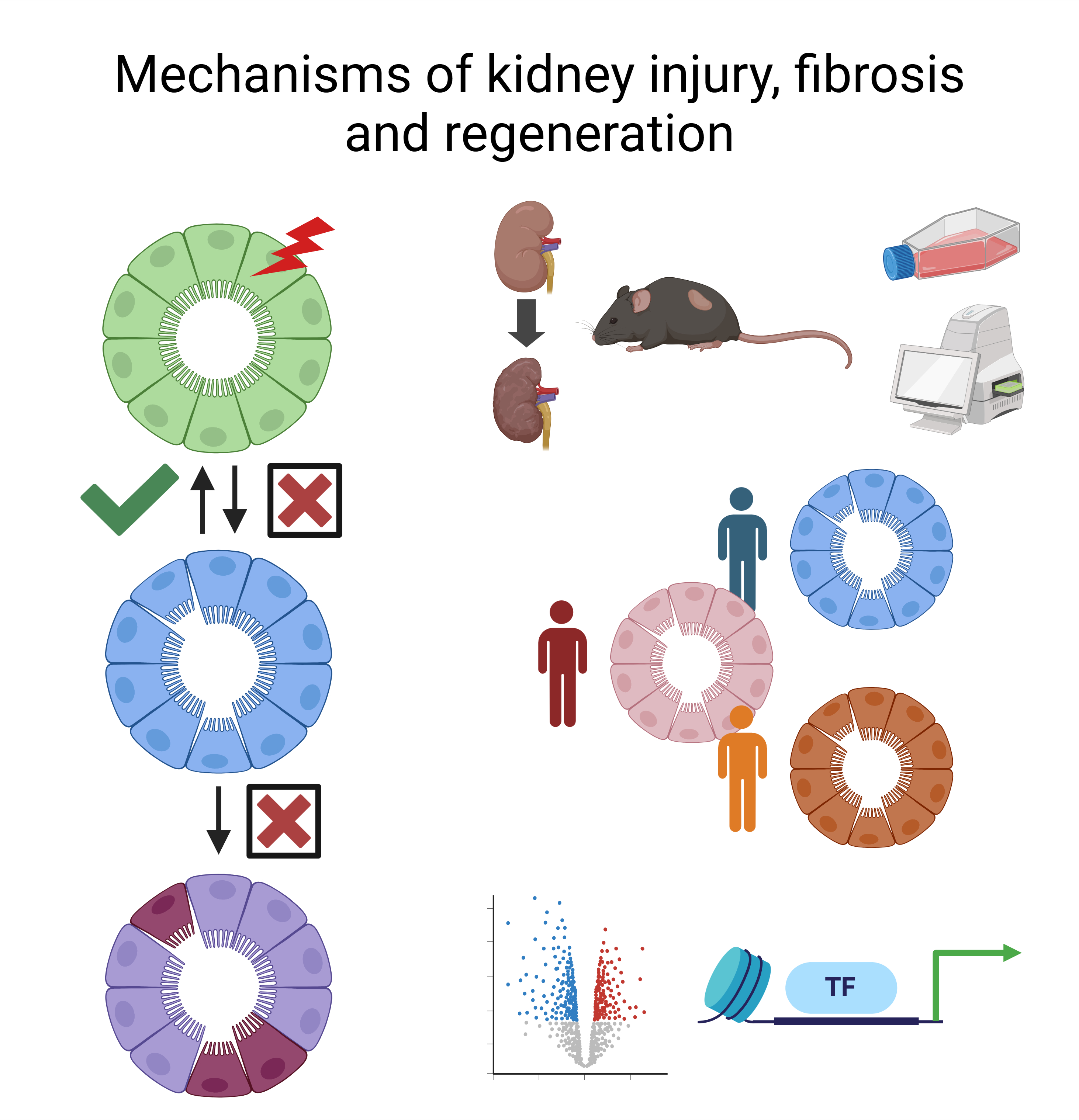
In the Li Lab, we study mechanisms of tissue fibrosis and primarily use kidney as the organ model, with the goal of identifying therapeutic targets that can ameliorate kidney disease progression. We specifically focus on kidney tubular injury and repair, metabolic dysfunction, dysregulated cell-cell communications and the underlying epigenetic mechanisms during acute kidney injury (AKI)-to-CKD transition.
In the past few years, we have developed or employed cutting-edge spatial and single-cell multiomics technologies to better understand these questions, including split-pool barcoding-based high-throughput single-cell sequencing (sci-RNA-seq; SHARE-seq), spatially resolved transcriptomics (DBiT-seq), epigenomics (spatial-ATAC-seq; spatial-CUT&Tag) and metabolomics (MALDI), as well as multimodal integration and analysis.
Kidney fibrosis represents the final common pathway of all chronic kidney diseases (CKD). Key challenges in addressing kidney fibrosis and halting CKD progression include understanding how to effectively target the health-to-disease transition of tubular epithelial cells and discovering strategies to promote cell redifferentiation and regeneration following kidney injury.
Our research integrates human clinical resources, mouse models of kidney injury and fibrosis, tissue culture systems, real-time metabolic measurements, multiomics and bioinformatics approaches to uncover the genetic and epigenetic mechanisms driving kidney diseases.
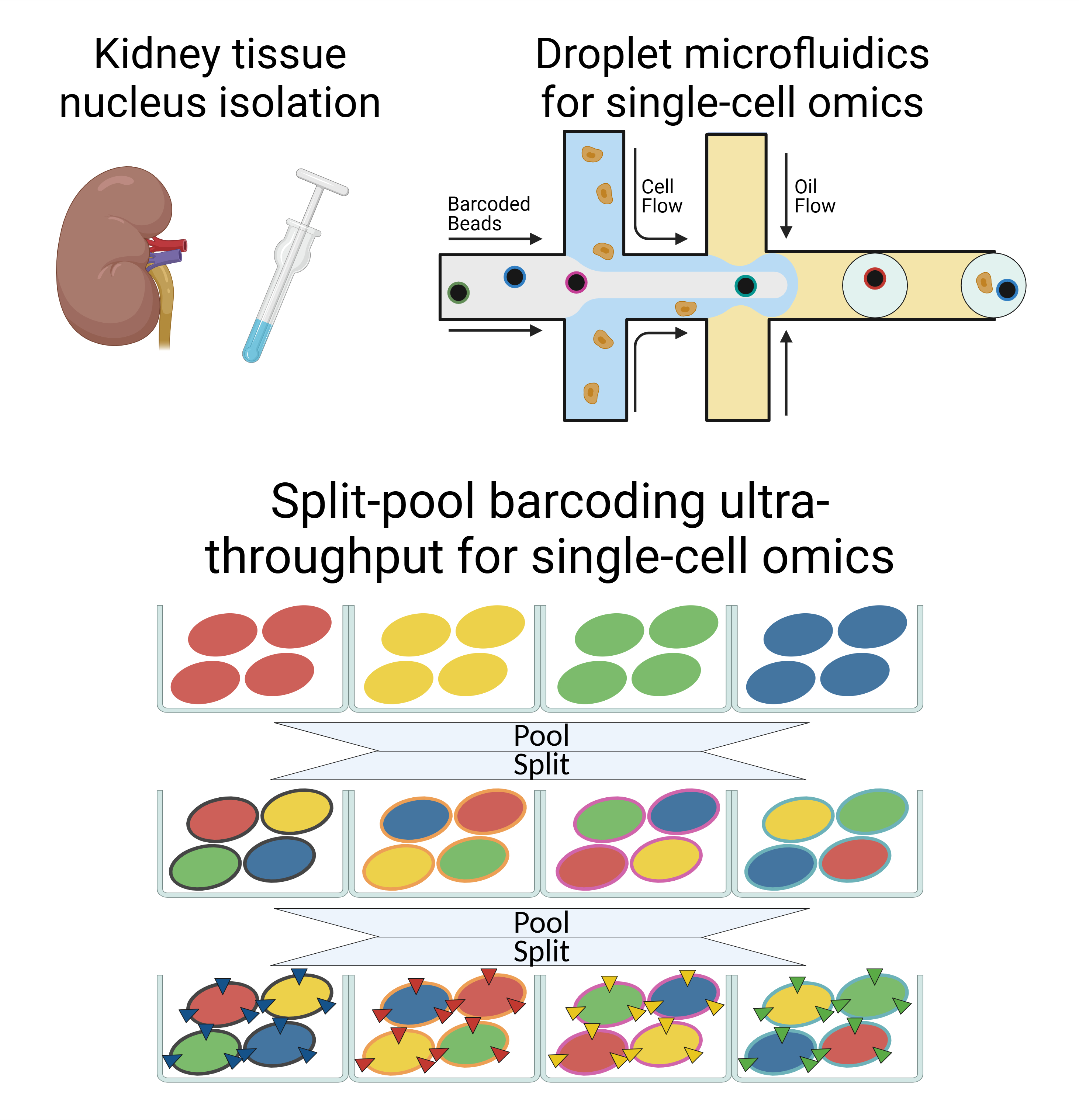
We have deep expertise in development and applications of various single-cell technologies. Beyond the well-known droplet microfluidics approach, we employ the split-pool barcoding method, which offers unique advantages in ultra-high throughput, robust sample multiplexing and cost efficiency.
The creation of the kidney single-cell transcriptomics reference atlas (Human Cell Atlas v1) marked a significant milestone. Now, we stand at the forefront of a transformative era, poised to develop a more comprehensive atlas — one characterized by greater coverage, a broader spectrum of disease subtypes, and enhanced multimodal complexity (HCA v2).
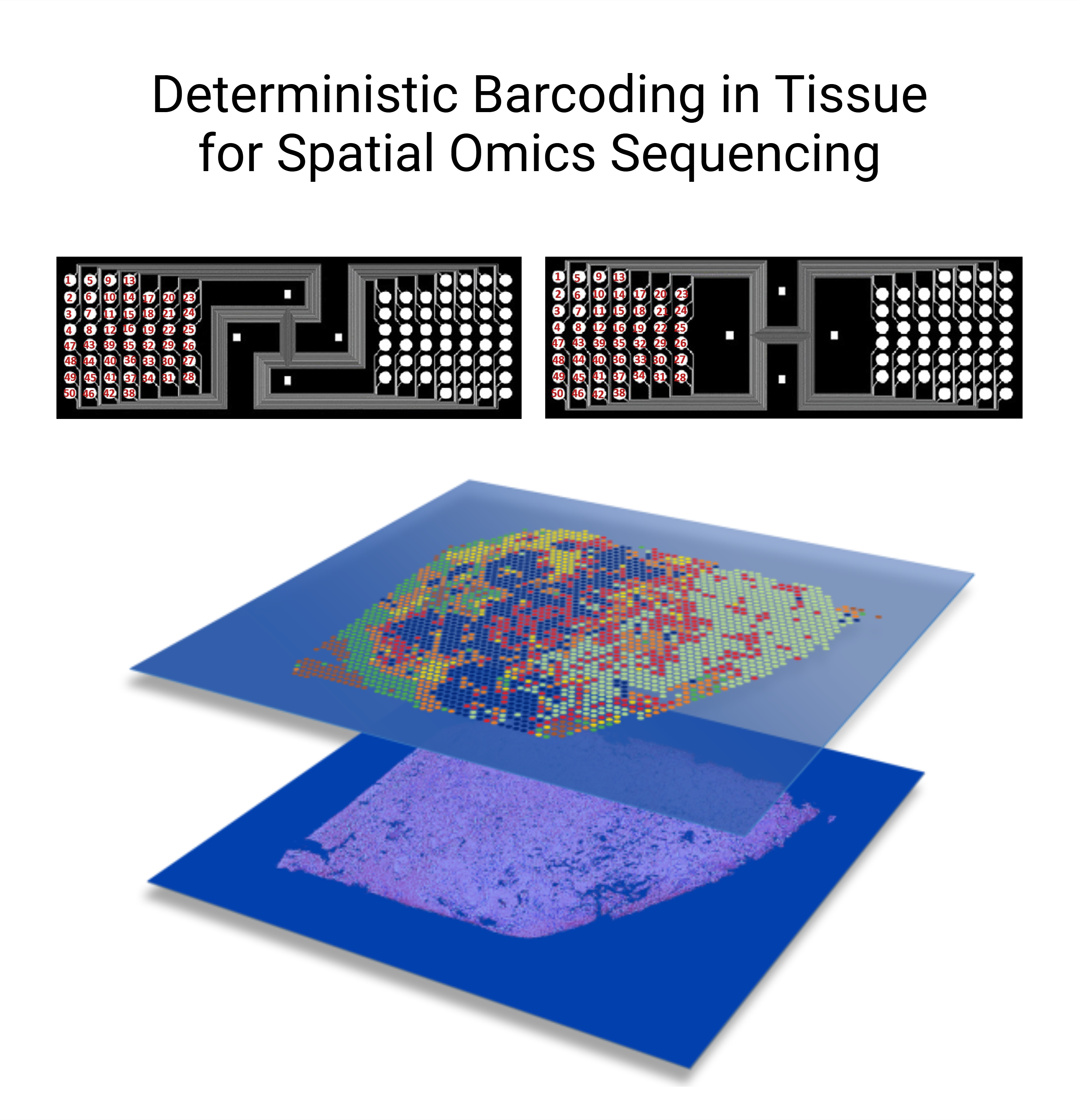
Never before have we had the ability to observe a tissue section at such a comprehensive molecular level and high resolution as we do today. The advent of spatial multiomics offers us invaluable opportunities to deepen our understanding of the structural changes in tissue organization and alterations in cell communication mechanisms underlying kidney physiology and disease states.
We are using cutting-edge Deterministic Barcoding in Tissue (DBiT) approaches to achieve near-single-cell resolution and whole-genome coverage for exploring spatial transcriptomics and spatial epigenomics. Furthermore, we aim to develop methods to enable spatial multimodal profiling and integrative analysis.